Data structure
Dataset structure¶
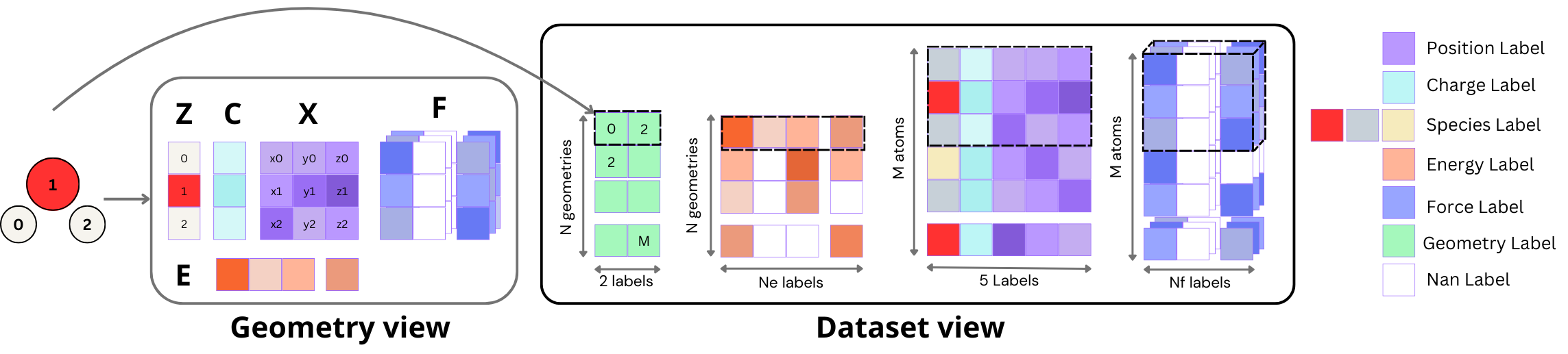
For a dataset with N geometries, M atoms across all geometries, ne energy labels, and nf force labels, we use zarr or memory-mapped arrays of various sizes:
- (M, 5) for atomic numbers (1), charges (1), and positions (3) of individual geometries;
- (N, 2) for the beginning and end indices of each geometry in the previous array;
- (N, ne) for the energy labels of each geometry, extendable to store other geometry-level QM properties such as HOMO-LUMO gap;
- (M, nf , 3) for the force labels of each geometry, extendable to store other atom-level QM properties.
The memory-mapped files efficiently access data stored on disk or in the cloud without reading them into memory, enabling training on machines with smaller RAM than the dataset size and accommodating concurrent reads in multi-GPU training. This allows for very efficient indexing, batching and iteration.
Formats¶
We currently support the following formats:
1) Zarr : https://zarr.readthedocs.io/en/stable/index.html
2) Memmap : https://numpy.org/doc/stable/index.html